.bg-text[ # Introduction to `ggplot2` ## Data Science for Biologists, Fall 2021 ] --- # Using R libraries/packages 1. **Install** the library/package one time. ```r install.packages("nameoflib") ``` 2. **Load** the library/package for *each R session/Rmd/script* where you want to use it. ```r library(nameoflib) ``` <br><br> **All packages you _need_ have been installed for you in RStudio Cloud Projects inside the Class Workspace.** You have to install them yourself in your Personal Workspace. --- # We are using the `tidyverse` packages .extra-large[ https://www.tidyverse.org/ ] --- # Loading the core tidyverse once installed: ```r #library(tidyverse) <-- without quotes also works here library("tidyverse") ``` ### If you don't load your package, you can't use its functions > Ok, only kind of. We'll talk more about this later! Example of an error: ``` Error in ggplot() : could not find function "ggplot" ``` --- <img src="img/ggplot2//ggplot2-hex.png" width="500px" /> --- # The dataset ```r msleep_smol ``` ``` ## # A tibble: 17 × 7 ## name vore order conservation awake brainwt bodywt ## <chr> <fct> <fct> <fct> <dbl> <dbl> <dbl> ## 1 Greater short-tailed shrew omni Soric… lc 9.1 0.00029 0.019 ## 2 Guinea pig herbi Roden… domesticated 14.6 0.0055 0.728 ## 3 Chinchilla herbi Roden… domesticated 11.5 0.0064 0.42 ## 4 Star-nosed mole omni Soric… lc 13.7 0.001 0.06 ## 5 Lesser short-tailed shrew omni Soric… lc 14.9 0.00014 0.005 ## 6 Long-nosed armadillo carni Cingu… lc 6.6 0.0108 3.5 ## 7 Tree hyrax herbi Hyrac… lc 18.7 0.0123 2.95 ## 8 North American Opossum omni Didel… lc 6 0.0063 1.7 ## 9 European hedgehog omni Erina… lc 13.9 0.0035 0.77 ## 10 Domestic cat carni Carni… domesticated 11.5 0.0256 3.3 ## 11 Gray hyrax herbi Hyrac… lc 17.7 0.0123 2.62 ## 12 Golden hamster herbi Roden… en 9.7 0.001 0.12 ## 13 House mouse herbi Roden… nt 11.5 0.0004 0.022 ## 14 Rabbit herbi Lagom… domesticated 15.6 0.0121 2.5 ## 15 Laboratory rat herbi Roden… lc 11 0.0019 0.32 ## 16 Arctic ground squirrel herbi Roden… lc 7.4 0.0057 0.92 ## 17 Thirteen-lined ground squirrel herbi Roden… lc 10.2 0.004 0.101 ``` --- # The dataset ```r str(msleep_smol) ``` ``` ## tibble [17 × 7] (S3: tbl_df/tbl/data.frame) ## $ name : chr [1:17] "Greater short-tailed shrew" "Guinea pig" "Chinchilla" "Star-nosed mole" ... ## $ vore : Factor w/ 3 levels "carni","herbi",..: 3 2 2 3 3 1 2 3 3 1 ... ## $ order : Factor w/ 13 levels "Afrosoricida",..: 13 11 11 13 13 3 7 4 6 2 ... ## $ conservation: Factor w/ 5 levels "domesticated",..: 3 1 1 3 3 3 3 3 3 1 ... ## $ awake : num [1:17] 9.1 14.6 11.5 13.7 14.9 6.6 18.7 6 13.9 11.5 ... ## $ brainwt : num [1:17] 0.00029 0.0055 0.0064 0.001 0.00014 0.0108 0.0123 0.0063 0.0035 0.0256 ... ## $ bodywt : num [1:17] 0.019 0.728 0.42 0.06 0.005 3.5 2.95 1.7 0.77 3.3 ... ``` --- # Let's dive in: Scatterplots in `ggplot2` ### Goal: Visualize the relationship between body weight (`bodywt`) and brain weight `brainwt`) of all smol mammals, where `bodywt` is across `brainwt` -- ### Step 1: *Roughly draw/plan the plot by hand. I am completely serious* --- # Scatterplots in `ggplot2` ```r ggplot(msleep_smol) ``` <!-- --> --- # Scatterplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = brainwt, y = bodywt) ``` <!-- --> ### `aes()` _MAPS_ columns (variables!) onto the plot. --- # Scatterplots in `ggplot2` ```r ggplot(msleep_smol) + (x = brainwt, y = bodywt) ``` ``` ## Error: <text>:2:15: unexpected ',' ## 1: ggplot(msleep_smol) + ## 2: (x = brainwt, ## ^ ``` ### Don't forget `aes()`! --- # Scatterplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = brainwt, y = bodywt) + * geom_point() ``` 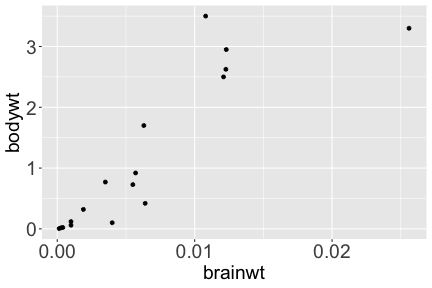<!-- --> --- # Scatterplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = brainwt, y = bodywt) + * geom_point(color = "firebrick") ``` 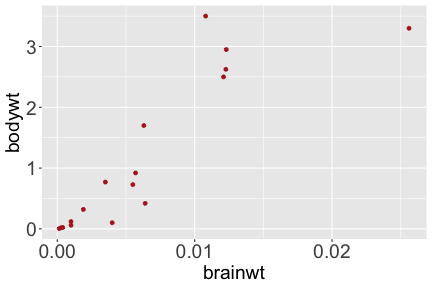<!-- --> --- # Scatterplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = brainwt, y = bodywt) + * geom_point(color = "firebrick", size = 3) ``` 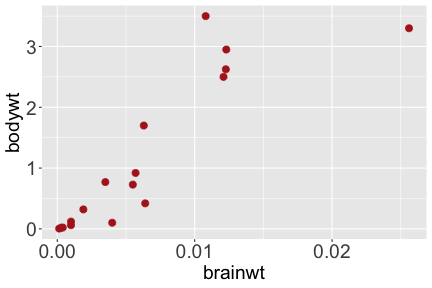<!-- --> --- # Plots can be saved just like variables ```r sleep_scatter <- ggplot(msleep_smol) + aes(x = brainwt, y = bodywt) + geom_point(color = "firebrick", size = 3) sleep_scatter ``` 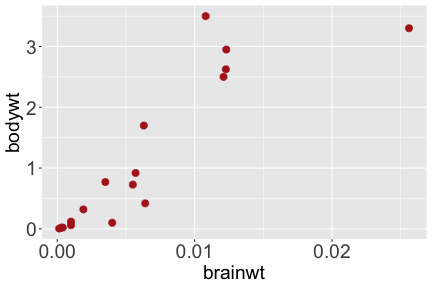<!-- --> --- # Don't forget the _forward_ assignment operator ```r ggplot(msleep_smol) + aes(x = brainwt, y = bodywt) + geom_point(color = "firebrick", size = 3) -> sleep_scatter sleep_scatter ``` 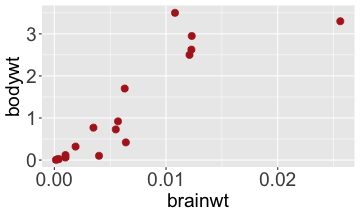<!-- --> --- # Adding onto an existing plot ```r ggplot(msleep_smol) + aes(x = brainwt, y = bodywt) + geom_point(color = "firebrick", size = 3) ``` ```r ggplot(msleep_smol) + aes(x = brainwt, y = bodywt) -> sleep_xy sleep_xy + geom_point(color = "firebrick", size = 3) ``` 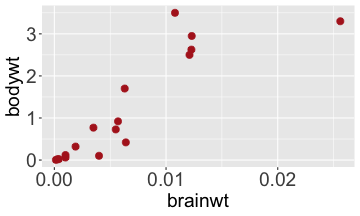<!-- --> --- # Quick tangent: The plot theme > The default theme is `theme_gray()` (`theme_grey()`) .pull-left[ ```r sleep_scatter ``` 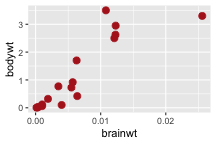<!-- --> ```r sleep_scatter + theme_classic() ``` <!-- --> ] .pull-left[ ```r sleep_scatter + theme_dark() ``` <!-- --> ```r sleep_scatter + theme_minimal() ``` <!-- --> ] --- # Adding on a theme ```r ggplot(msleep_smol) + aes(x = brainwt, y = bodywt) + geom_point() + * theme_bw() ``` <!-- --> --- # Setting a theme for _all plots_ All plots created _after_ this line is run will use `theme_classic()`. All remaining plots in the slides will now use this theme as the new default. <br><br> ```r theme_set(theme_classic()) ``` + `get_help("theme_set")` + `get_help("theme_classic")` + All the built-in themes in action: https://spielmanlab.github.io/introverse/introverse_docs/ggplot2_theme_gray.html + Customizing themes: https://spielmanlab.github.io/introverse/articles/themes.html --- # Histograms in `ggplot2` ### Goal: Visualize the distribution of all mammal times spent awake (`awake`) -- ### Step 1: Plan the plot *by hand* --- # Histograms in `ggplot2` -- ```r ggplot(msleep_smol) ``` <!-- --> --- # Histograms in `ggplot2` ```r ggplot(msleep_smol) + * aes(x = awake) ``` <!-- --> --- # Histograms in `ggplot2` ```r ggplot(msleep_smol) + aes(x = awake) + * geom_histogram() ``` ``` ## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`. ``` <!-- --> --- # Histograms in `ggplot2` ```r ggplot(msleep_smol) + aes(x = awake) + * geom_histogram(bins = 10) ``` <!-- --> ### Use the argument `bins` to specify literally how many bins there should be --- # Histograms in `ggplot2` ```r ggplot(msleep_smol) + aes(x = awake) + * geom_histogram(binwidth = 2) ``` <!-- --> ### OR, use the argument `binwidth` to specify how wide the bins should be along the X-axis --- # "Trial and error" to find the "right" binning .pull-left[ ```r ggplot(msleep_smol) + aes(x = awake) + * geom_histogram(bins = 2) ``` <!-- --> ] .pull-right[ ```r ggplot(msleep_smol) + aes(x = awake) + * geom_histogram(bins = 30) ``` <!-- --> ] --- # Histograms in `ggplot2` ```r ggplot(msleep_smol) + aes(x = awake) + * geom_histogram(binwidth = 2, color = "dodgerblue") ``` <!-- --> --- # Histograms in `ggplot2` ```r ggplot(msleep_smol) + aes(x = awake) + * geom_histogram(binwidth = 2, * color = "dodgerblue") ``` <!-- --> ### When code gets long, separate onto multiple lines for clarity. _YOU WILL BE GRADED ON THIS!!!!_ --- # Histograms in `ggplot2` ```r ggplot(msleep_smol) + aes(x = awake) + geom_histogram(binwidth = 2, color = "dodgerblue", * fill = "orange") ``` <!-- --> --- # Histograms in `ggplot2` ```r ggplot(msleep_smol) + aes(x = awake) + geom_histogram(binwidth = 2, color = "dodgerblue", fill = "orange", * size = 3) ``` <!-- --> <br> **This is _pretty ugly_, but now you've learned about `size`!** --- # COMMENT COMMENT COMMENT COMMENT ```r ggplot(msleep_smol) + # specify x on the x-axis aes(x = awake) + geom_histogram(binwidth = 2, # use binwidth of 2 # outline the histogram in blue color = "dodgerblue", # fill the bars with orange fill = "orange", # increase width of histogram lines size = 3) ``` <!-- --> --- # Density plots in `ggplot2` ### Goal: Visualize the distribution of all mammal times spent awake (`awake`) for each vore -- ### Step 1: Draw it by hand --- # Density plots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = awake) + * geom_density() ``` <!-- --> --- # Density plots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = awake) + * geom_density(fill = "cornflowerblue", * color = "chartreuse", * size = 4) ``` <!-- --> ### Again, kinda ugly, but look how easy that was to make!! --- # What if we wanted to show *all vores* separately? -- ### Conceptualize by hand: --- # Density plots in `ggplot2` ```r ggplot(msleep_smol) + * aes(x = awake, fill = vore) + geom_density() ``` 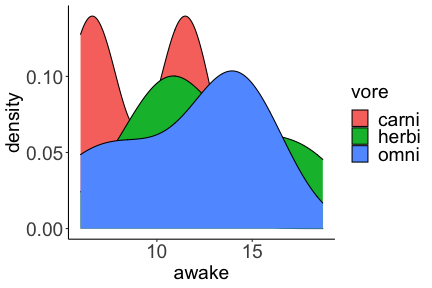<!-- --> ### We need to make the density plots _transparent_. --- # Density plots in `ggplot2` ```r ggplot(msleep_smol) + * aes(x = awake, fill = vore) + * geom_density(alpha = 0.5) ``` 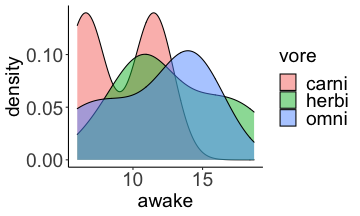<!-- --> + `alpha` is NOT a variable, so it does NOT belong in `aes()`. It is a visual aspect of the *density plot*, so we provide the argument to `geom_density()` + `alpha = 1`: Completely transparent. + `alpha = 0`: Completely opaque --- # Density plots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = awake, * fill = vore, * color = vore) + geom_density(alpha = 0.5) ``` 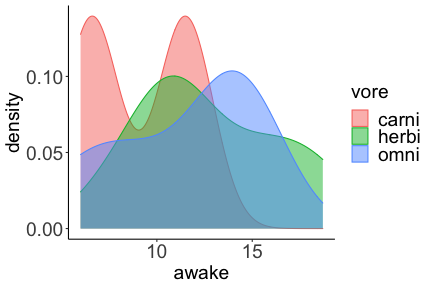<!-- --> ### We will learn how to customize these colors next week --- # Boxplots in `ggplot2` ### Goal: Visualize the distribution of all mammal times spent awake (`awake`) for each vore ### Step 1: --- # Boxplots in `ggplot2` ```r ggplot(msleep_smol) + * aes(x = vore, y = awake) ``` <!-- --> --- # Boxplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore, y = awake) + * geom_boxplot() ``` 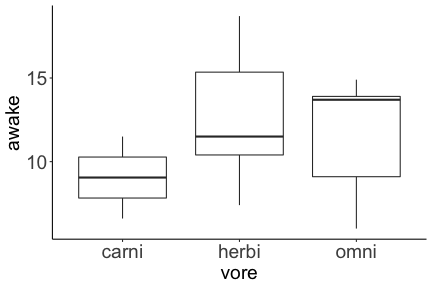<!-- --> --- # Boxplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore, y = awake) + * geom_boxplot(color = "deeppink", * fill = "goldenrod1") ``` 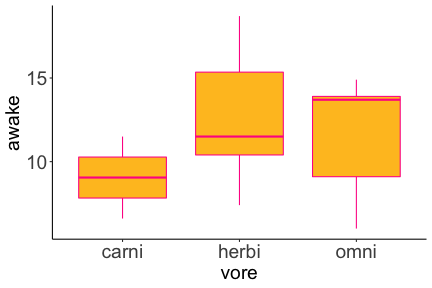<!-- --> -- ### What if we want a separate fill _for each vore_? --- # Boxplots in `ggplot2` ```r ggplot(msleep_smol) + * aes(x = vore, y = awake, fill = vore) + geom_boxplot() ``` 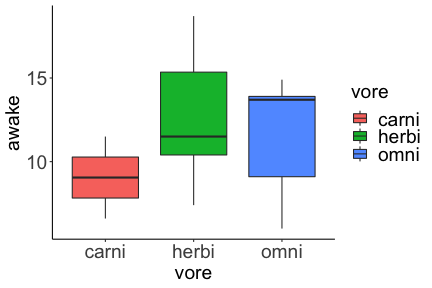<!-- --> --- # Violin plots in `ggplot2` .pull-left[ ```r ggplot(msleep_smol) + aes(x = vore, y = awake) + * geom_violin() ``` <!-- --> ] .pull-right[ ```r ggplot(msleep_smol) + aes(x = vore, y = awake, * fill = vore) + geom_violin() ``` <!-- --> ] --- # Strip/jitter plots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore, y = awake) + # size = 2 to see more easily in slides * geom_jitter(size = 2) ``` <!-- --> --- # Strip/jitter plots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore, y = awake) + * geom_jitter(width = 0.2, size = 2) ``` <!-- --> ### Usually need to pick a width between 0.1 - 0.3, *in my opinion* --- # Strip/jitter plots in `ggplot2` ```r ggplot(msleep_smol) + # COLOR, NOT FILL!! These are points aes(x = vore, y = awake, * color = vore) + geom_jitter(width = 0.2, size = 2) ``` 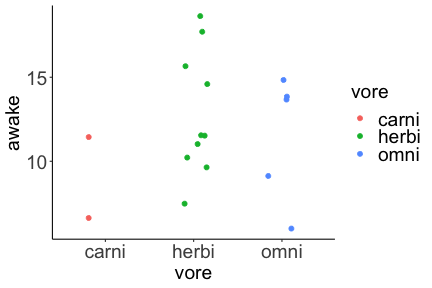<!-- --> <br> **Did you notice the points were placed little differently each plot version?** --- # Other types of points + Any time you use `geom_point()` or `geom_jitter()` you can specify a different point! + _Point shapes 21-25 have a color and a fill_ <img src="img/ggplot2//r_pch.png" width="350px" /> --- # Example of shape 21 ```r ggplot(msleep_smol) + aes(x = vore, y = awake, * fill = vore) + geom_jitter(width = 0.2, size = 4, * color = "black", * shape = 21) ``` 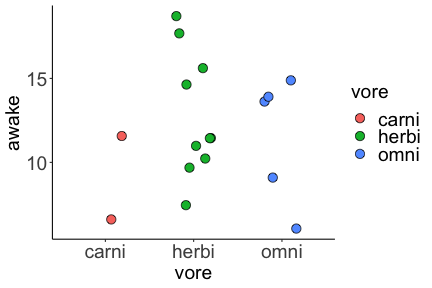<!-- --> --- # Barplots in `ggplot2` ### Goal: How many of each vore are there? > Note: There are many ways to make barplots. Today we will learn how to make barplots specifically to show _amount of a categorical variable._ ### Step 1: --- # Barplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore) + * geom_bar() ``` 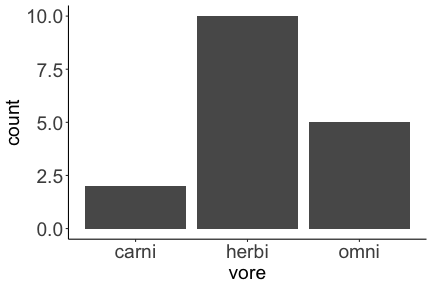<!-- --> --- # Barplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore) + * geom_bar(fill = "blueviolet") ``` 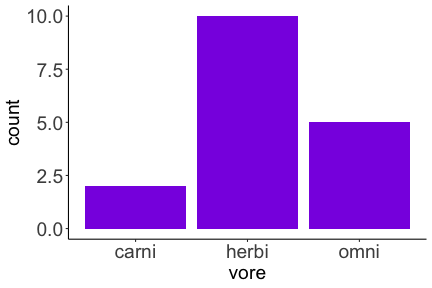<!-- --> --- # Barplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore) + * geom_bar(fill = "blueviolet", * color = "black", size = 2) ``` <!-- --> --- # Barplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore, * fill = vore) + geom_bar() ``` <!-- --> --- # Barplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore, fill = vore) + * geom_bar(color = "black") ``` <!-- --> --- # Barplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore, * fill = conservation) + geom_bar() ``` <!-- --> --- # Barplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore, fill = conservation) + * geom_bar(position = position_dodge()) ``` <!-- --> --- # Barplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore, fill = conservation) + * geom_bar(position = position_dodge(preserve="single")) ``` <!-- --> --- # Barplots in `ggplot2` .pull-left[ ```r ggplot(msleep_smol) + aes(x = vore, fill = conservation) + geom_bar() ``` <!-- --> ] .pull-right[ ```r ggplot(msleep_smol) + aes(x = conservation, fill = vore) + geom_bar() ``` <!-- --> ] --- # Barplots in `ggplot2` ```r ggplot(msleep_smol) + aes(x = vore, fill = order) + geom_bar() ``` 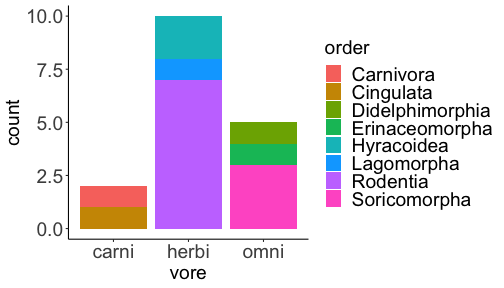<!-- --> ### My first impression of this plot is _feeling overwhelmed_. --- # Labeling your plots ```r ggplot(msleep_smol) + aes(x = vore) + geom_bar() + * labs(x = "X-axis label", * y = "Y-axis label", * title = "Plot title", * subtitle = "In case you wanted a subtitle", * caption = "Would you like a caption?") ``` 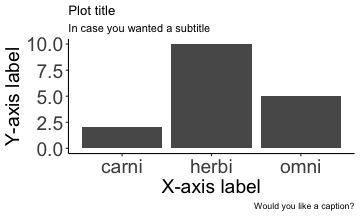<!-- --> Always make sure your x and y axes are professionally labeled (*no underscores!*). Title, subtitle, caption are optional to include. --- # Plots we can make! 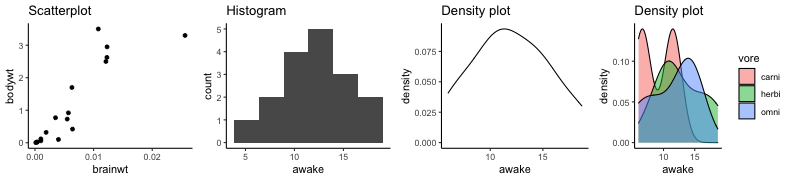<!-- -->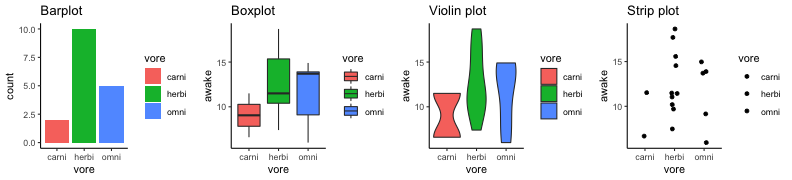<!-- --> --- # Adding panels ("facets") to plots ```r ggplot(msleep_smol, aes(x = awake)) + geom_histogram(binwidth = 2, color = "black", fill = "firebrick2") ``` 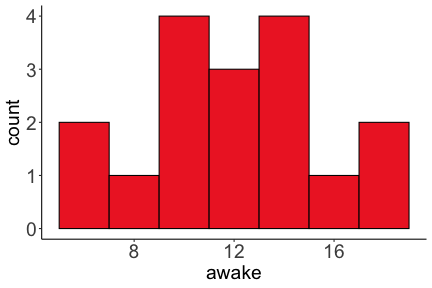<!-- --> ### What if we want a separate histogram for each `vore`? --- # This is a bad idea. ```r ggplot(msleep_smol, aes(x = awake, fill = vore)) + geom_histogram(binwidth = 2, color = "black") ``` 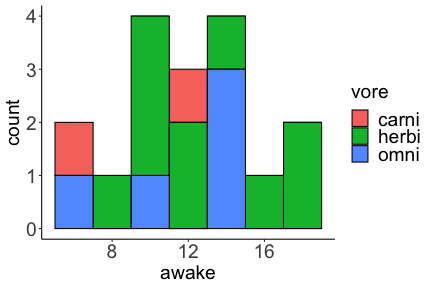<!-- --> --- # Adding panels ("facets") to plots ```r ggplot(msleep_smol, aes(x = awake)) + geom_histogram(binwidth = 2, # may need to tweak again! color = "black", fill = "firebrick2") + * facet_wrap( vars(vore) ) ``` 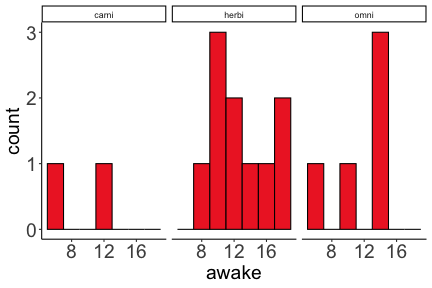<!-- --> <br> **Faceting is the _ONLY TIME_ we do not use `aes()` to refer to a column!!! We use `vars()` when faceting.** --- # Adding panels ("facets") to plots ```r ggplot(msleep_smol, aes(x = awake)) + geom_histogram(binwidth = 2, # may need to tweak again! color = "black", fill = "firebrick2") + # you get to choose which is row and which is column * facet_grid( rows = vars(vore), * cols = vars(conservation) ) ``` 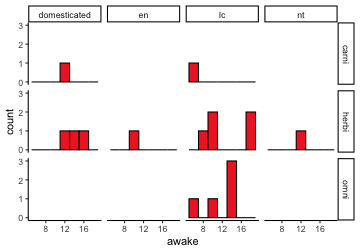<!-- --> **Use `facet_grid()` to make a panel grid across 2 variables. This plot is _pretty bad_, but it teaches `facet_grid()`!**