Model Selection and Validation
Lecture
R content
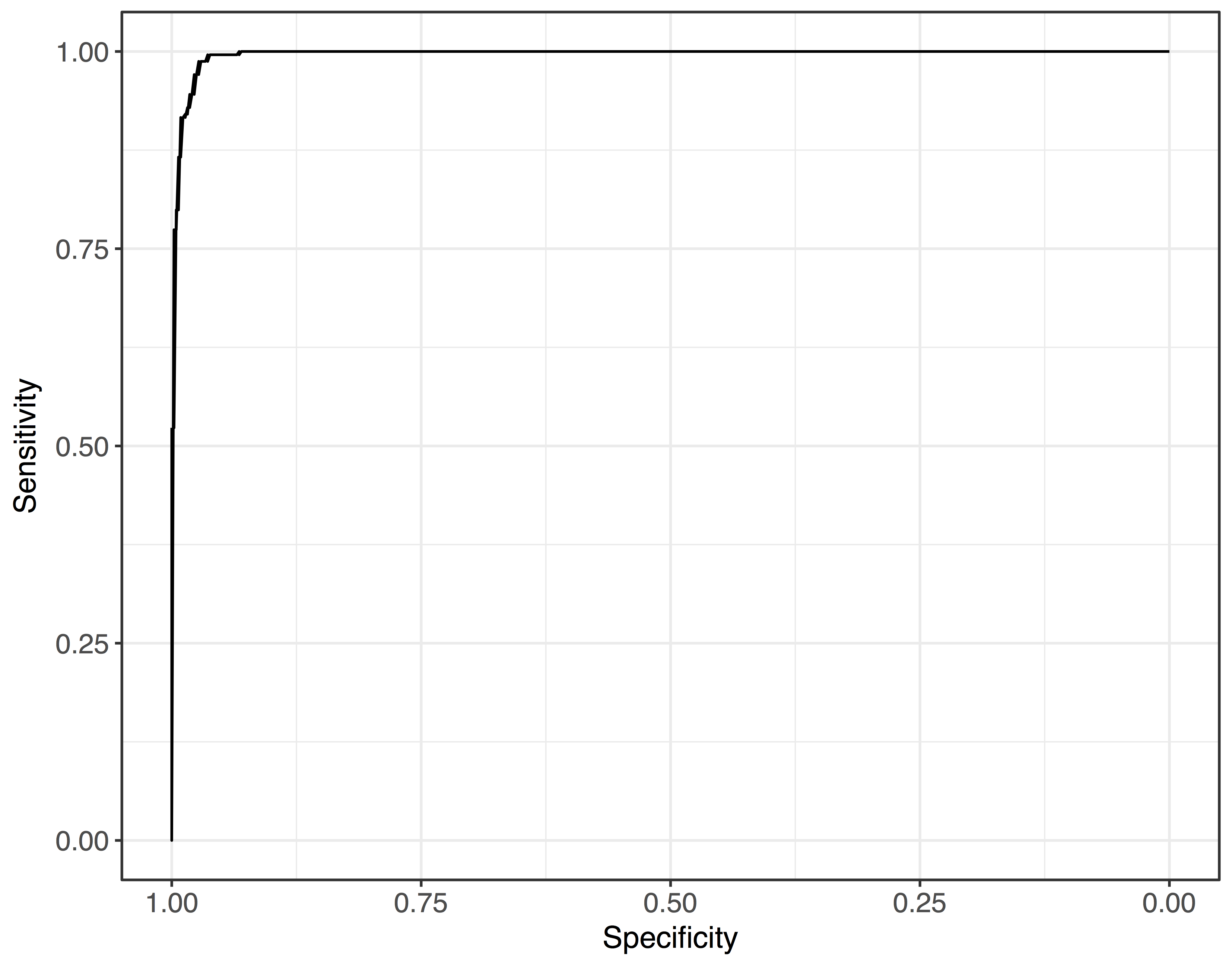
ROC curves and AUC
> library(pROC)
## fit a logistic regression to biopsy data
> model <- glm(outcome ~ ., data=biopsy, family=binomial)
## create ROC object
> roc.object <- roc(biopsy$outcome, model$linear.predictors)
## extract AUC
> roc.object$auc
Area under the curve: 0.9963
## Plot!
> roc.data <- tibble(x = roc.object$specificities, ### TNR (1-FPR)
y = roc.object$sensitivities) ### TPR
> ggplot(roc.data, aes(x = x, y = y)) +
geom_line() + scale_x_reverse() +
ylab("Sensitivity") +
xlab("Specificity")
Likelihood ratio test
Used to compare nested models.
> null_model <- lm(Sepal.Length ~ Petal.Length, data = iris)
> null_glance <- glance(null_model)
> alt_model <- lm(Sepal.Length ~ Petal.Length + Species, data = iris)
> alt_glance <- glance(alt_model)
#### LRT #####
> D <- 2 * (alt_glance$logLik - null_glance$logLik) ### test statistic
> df <- alt_glance$df - null_glance$df ### chisquared degrees of freedom
> 1 - pchisq(D,df)
[1] 2.799982468e-13
With P=2.79e-13, we have evidence in favor of the alternative model, i.e. with the Species predictor added in.
Stepwise model selection
The step() function in base R is one of many options for step-wise model selection. By default, it uses AIC but you can change with the criterion argument. The output of this function is the selected model itself.
> model <- lm(Sepal.Length ~ ., data = iris)
### Selection with AIC
> aic.backwards <- step(model, trace=F) ## trace=F (or trace=0) reduces output vomit
# To prove to you that the output is a model:
> glance(aic.backwards)
> tidy(aic.backwards)
term estimate std.error statistic p.value
1 (Intercept) 2.1712663 0.27979415 7.760227 1.429502e-12
2 Sepal.Width 0.4958889 0.08606992 5.761466 4.867516e-08
3 Petal.Length 0.8292439 0.06852765 12.100867 1.073592e-23
4 Petal.Width -0.3151552 0.15119575 -2.084418 3.888826e-02
5 Speciesversicolor -0.7235620 0.24016894 -3.012721 3.059634e-03
6 Speciesvirginica -1.0234978 0.33372630 -3.066878 2.584344e-03
#### Selection with BIC, for example
> bic.backwards <- step(model, trace=F, criterion = "BIC")
>